For those of you that follow the updates of GA4GH you will have seen the unanimous approval of the steering committee for DUO to be included in its suite of technical standards for the sharing of genomic and health related data.
DUO has three main features:
- Each term has been generated by the community and includes a human readable definition that can be expanded where necessary.
- It is a machine-readable file that encodes how that data can be used and how a researcher intends to use the data.
- Can be implemented alongside an advanced search algorithm which would allow authenticated users to query and gain access to datasets pertaining to their research. e.g., an industry researcher working on cancer could potentially be matched to any dataset that is allowed for commercial use and for cancer research and offered the opportunity to fetch them automatically.
So, what does this mean for EGA? EGA as a driver project has adopted this standard and will be utilizing it for two main purposes in the first instance.
i) It will allow EGA users to instantly identify what terms they will need to
agree to as part of any Data Access Agreements. This will save valuable time
for both applicants and Data Access Committees alike - as you will be able to
see if you are likely to be able to access the data based on your research
intentions and working background.
ii) In the future it will allow users to be able to search for data based on
Data Use Ontology terms e.g., you could search for all data that can be used
for
General Research Use (GRU).
As EGA moves forwards submitters will be asked to align their data access policies with DUO so that their dataset(s) can be tagged appropriately on our web pages.
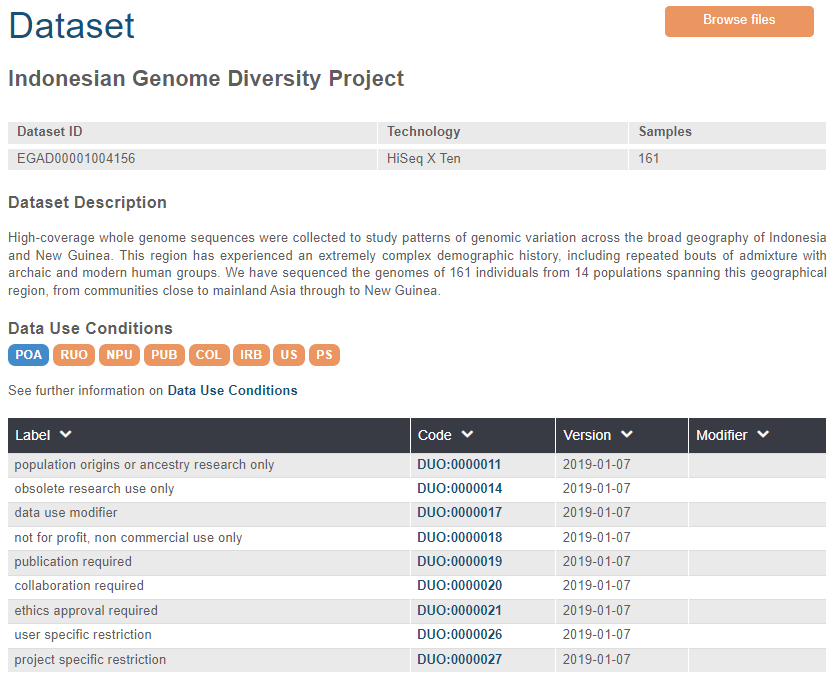
If you would like to add DUO to any of your existing datasets, or to any future datasets, do please just let us know.