Cancer in children is uncommon and the overall prognosis for most pediatric cancers is good. However, while the combined survival rates have improved over the last decades, certain childhood malignancies, such as high-grade gliomas or metastatic sarcomas, still remain incurable in most patients. New strategies for targeting those devastating diseases are imperative, and patient genomic data can become a key asset in the process [1].
The Pediatric Cancer Genome Project’s datasets
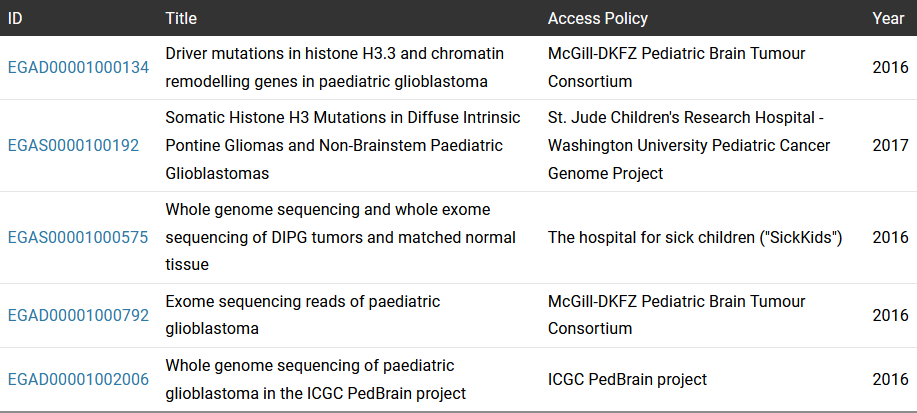
At EGA, we store over 500 datasets with sequencing data from pediatric cancer patients. A remarkable case is the datasets belonging to the Pediatric Cancer Genome Project (PCGP) from St. Jude Children’s Research Hospital–Washington University (Table 1 - EGAS list). PCGP is an ambitious effort to identify the mutations that drive childhood cancer and find new cures. Those datasets include 600 patients with complete tumor and normal genomes from 15 different tumor types. PCGP datasets have been an important resource for various studies that pooled genomic data from different public and in-house datasets to perform extensive genomic characterizations and publish comprehensive pan-cancer pediatric studies [2],[3]. Other interesting pediatric cancer datasets are from the Pediatric Brain Tumor Consortium, Sickkids or ICGC PedBrain project.
How data reuse impacts drug development for pediatric cancers
Recently, in October 2024, Nature Communications published a work led by the University of Michigan where authors used data from EGA (PCGP) check the table below as part of their strategy to pursue the identification of new tumor vulnerabilities susceptible to becoming novel therapeutic targets in diffuse midline glioma. Diffuse midline gliomas (DMG) are treatment-resistant and uniformly fatal pediatric brain tumors. The prognosis of this brainstem tumor is dismal with a median overall survival of 9–12 months from diagnosis.
In this study, the authors first developed a lab model of the disease and reanalyzed data from the EGA to identify genes potentially linked to tumor severity. This approach revealed involvement of specific metabolic pathways, suggesting new possibilities for therapeutic intervention. In mice, the study shows that the use of statins can improve survival.
For a brief overview of the underlying science, diffuse midline gliomas present intratumor heterogeneity with subpopulations of less-differentiated oligodendrocyte precursors and more differentiated astrocytes. Authors established in vitro models to recapitulate both phenotypes and identify metabolic programs in both subpopulations. To determine the clinical relevance, the authors re-used gene expression data from 76 DMG patients identifying a gene signature predicting decreased overall survival. After extensive metabolic characterization of subpopulations, authors defined strategies to target specific metabolic vulnerabilities. Pre-clinical experiments in mice using OXPHOS inhibitors and statins showed a reduction in tumor burden and increased overall survival [4].
In this scenario, the re-use of high-quality patient’s sequencing data appeared as an advantageous strategy to support the clinical relevance of in-vitro and pre-clinical findings. On the other hand, in rare conditions such as childhood cancer, the availability of public datasets and the possibility of pooling data from different sources can be the only way to obtain impactful results that lead to improvements for patients.
References
- Davidoff, A. M. Pediatric Oncology. Seminars in pediatric surgery 19, 225–233 (2010).
- Gröbner, S. N. et al. The landscape of genomic alterations across childhood cancers. Nature 555, 321–327 (2018).
- Venu Thatikonda et al. Comprehensive analysis of mutational signatures reveals distinct patterns and molecular processes across 27 pediatric cancers. Nature Cancer 4, 276–289 (2023).
- Mbah, N. E. et al. Therapeutic targeting of differentiation-state dependent metabolic vulnerabilities in diffuse midline glioma. Nature Communications 15, (2024).
Datasets and Studies
| ID | Title | Access Policy | Year |
|---|---|---|---|
| EGAD00001000134 | Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma | McGill-DKFZ Pediatric Brain Tumour Consortium | 2016 |
| EGAS0000100192 | Somatic Histone H3 Mutations in Diffuse Intrinsic Pontine Gliomas and Non-Brainstem Paediatric Glioblastomas | St. Jude Children's Research Hospital - Washington University Pediatric Cancer Genome Project | 2017 |
| EGAS00001000575 | Whole genome sequencing and whole exome sequencing of DIPG tumors and matched normal tissue | The hospital for sick children ("SickKids") | 2016 |
| EGAD00001000792 | Exome sequencing reads of paediatric glioblastoma | McGill-DKFZ Pediatric Brain Tumour Consortium | 2016 |
| EGAD00001002006 | Whole genome sequencing of paediatric glioblastoma in the ICGC PedBrain project | ICGC PedBrain project | 2016 |