response efforts
On 11 March 2020 the World Health Organization declared the novel coronavirus outbreak a global pandemic. Four months later, the European Genome-phenome Archive (EGA) released its first COVID-19 dataset. This dataset – single cell RNA and VDJ sequencing of B cells from 60 COVID-19 patients – showed that neutralizing antibodies could be identified by high-throughput sequencing in response to SARS-CoV-2 infection. That was one year ago.
Today, the EGA provides access to fifteen COVID-19 datasets from researchers across seven countries in Asia, Europe, and North America. These studies represent almost 17,000 individuals and have resulted in at least sixteen publications and preprints.
Researchers deposit controlled access COVID-19 data at EGA
The global research community has come together rapidly to investigate the SARS-CoV-2 coronavirus and better understand the related disease, COVID-19. These research efforts generate valuable genetic and phenotypic data from patients and research participants that can be shared with approved researchers. The EGA enables sharing of this research data by providing a service for archiving and controlled distribution of sensitive data. Over the past year, the EGA has worked with researchers to archive and distribute COVID-19 data from high-throughput sequencing experiments, genotyping studies, and phenotypic information. These datasets investigate the immune system, blood, and cells and tissues of the lung, which are relevant for studying a contagious respiratory illness caused by a viral infection.
*Study Spotlight. In January 2021, Ancestry.com demonstrated the utility of deep phenotyping based on self-reported outcomes from a large population of mild and asymptomatic COVID-19 cases. They identified genetic associations with eight COVID-19 phenotypes and showed distinct patterns of association, most notably related to the chr3/SLC6A20/LZTFL1 and chr9/ABO regions. The supporting data is available at the EGA to approved researchers and includes both genotype and phenotype data for 15,000 individuals.
EGA collaborates with global COVID-19 community
Since the coronavirus outbreak, the EGA has collaborated with other life science resources to support discovery and access to COVID-19 datasets.
COVID-19 Host Genetics Initiative. With the NHGRI’s Genomic Data Science Analysis, Visualization, and Informatics Lab-space (AnVIL) platform, the EGA enables sharing of individual-level genetic and phenotypic data from the COVID-19 Host Genetics Initiative (HGI). This initiative aims to generate, share, and analyze data from COVID-19 host genetics research projects to better understand the genetic determinants of COVID-19 susceptibility, severity, and outcomes. In response to COVID-19, the EGA actively supports COVID-19 data submissions and integration of data access and flow into the COVID-19-HGI analysis platform.
*Study Spotlight. The COVID-19 HGI has combined individual-level data for 13,868 COVID-19 positive patients (N=7,167 hospitalized) from 17 cohorts in nine countries. The data were used to assess the association of the major common COVID-19 genetic risk factor (chromosome 3 locus tagged by rs10490770) with mortality, COVID-19-related complications, and laboratory values. The genotype and phenotype data for 10 of these cohorts is available at the EGA to approved researchers under accession EGAS00001005304.
European COVID-19 Data Portal. EGA-archived COVID-19 data are discoverable via the European COVID-19 Data Portal (Fig. 1), which brings together public and controlled access data to accelerate coronavirus research for the international community. By indexing all COVID-19-related data in one place, researchers can more easily discover relevant datasets of interest, thus increasing the “FAIRness” (Findability, Accessibility, Interoperability, Reusability) of this valuable data.
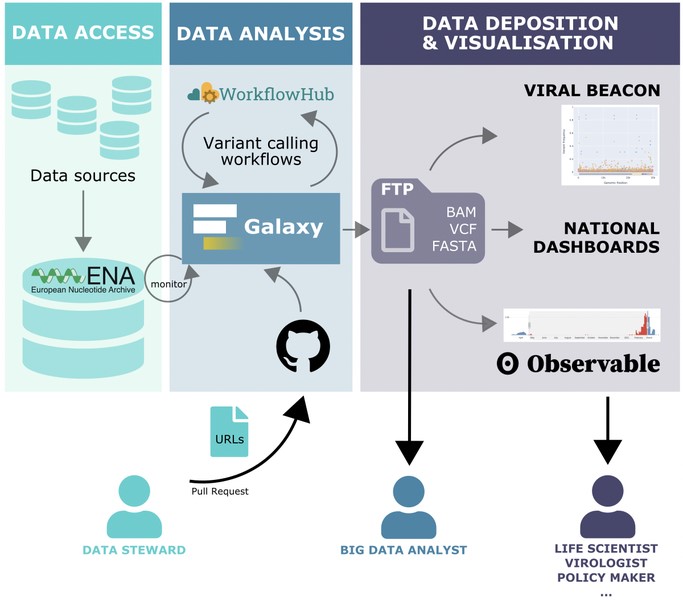
COVID-19 Viral Beacon. The COVID-19 Viral Beacon tool was developed in collaboration with the European Nucleotide Archive and Galaxy to enable near real-time browsing of SARS-CoV-2 variability at genomic, amino acid, and motif levels (Fig. 2). The COVID-19 Viral Beacon allows researchers to (i) perform detailed searches about genomic variants, (ii) filter queries and find unique cases, (iii) filter data based on strain-specific variants, and (iv) explore associated metadata. It uses the Global Alliance for Genomics and Health (GA4GH) Beacon standard including new Beacon v2 features. With this tool, researchers can study intra-host mutations on genomic regions of interest, or trace any variant frequency over time using raw read data. More than 200,000 SARS-CoV-2 analysed genomic data files are now available to researchers for further exploration.
Ongoing COVID-19 efforts at EGA
Addressing the COVID-19 pandemic is a global effort. Federated resources are necessary to support transnational deposition, access, and analysis of sensitive COVID-19 host genetics and other related data. At the same time, many countries now have emerging personalized medicine programmes which are generating data from national or regional healthcare initiatives. These data are subject to more stringent information governance than research data and often must comply with national data protection legislation. To address this need, the Federated EGA was established to serve as the primary global resource for discovery and access of sensitive human omics and associated data consented for secondary use. The Federated EGA will comprise a network of national human data repositories and will implement community standards and common interfaces.
Launching Federated EGA promises to accelerate not only global research efforts to understand, diagnose, and treat COVID-19, but also to foster data reuse, enable reproducibility, and accelerate biomedical and disease research to ultimately improve human health.



