CINECA, EUCANCan, and euCanSHare were part of the EUCAN Cluster, made up of six projects that received funding under the same Horizon 2020 call. All EUCAN projects (CINECA, EUCANCan, EUCAN-Connect, euCanSHare, Receptor Plus, and ReCoDID) were aimed at facilitating data reuse and knowledge discovery by enhancing data exchange and long-term collaboration in the health field.
Here are some highlights about EGA’s contribution to these projects.
CINECA: advances in Federated discovery and infrastructure for cohorts
CINECA (Common Infrastructure for National Cohorts in Europe, Canada, and Africa) developed a federated cloud-based infrastructure for making genomic and biomolecular data accessible. The project has assembled a virtual cohort of 1.4 million individuals from sources such as the EGA, CanDIG and H3Africa. The EGA–CRG co-leaded a work package Work Package 1 on Federated Data Discovery and Querying.
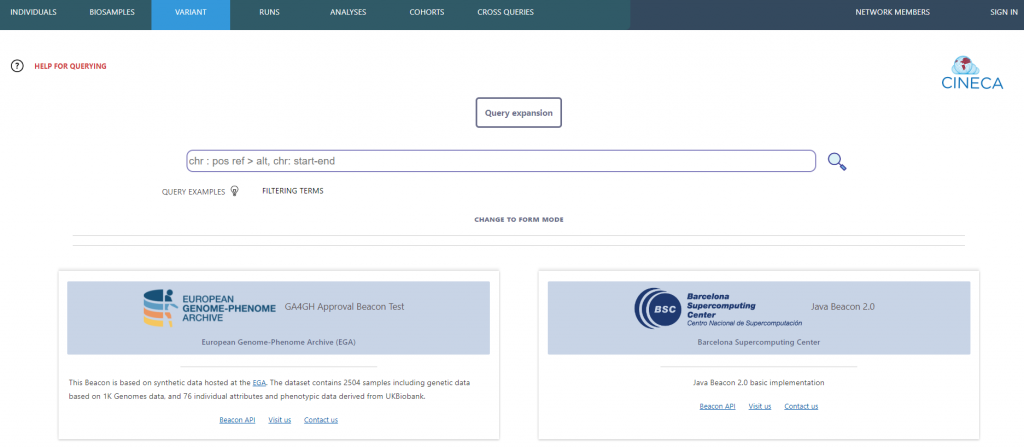
Beacon v2, championed by Jordi Rambla and Lauren Fromont, has been one of the central elements for the discovery of human genetic and phenotypic data. The EGA-CRG contributed to the development of a model for cohort discovery inside the Beacon v2 model. The team also delivered a Discovery Portal was implemented to explore cohorts and individuals of synthetic data. It is a UI that gathers a network of Beacons, a service for query expansion, and a visualisation tool.
euCanSHare: facilitation of data access management
This joint EU-Canada project aimed to build a European and Canadian FAIR platform for cardiovascular data sharing and analysis. The EGA’s tasks contributed to the development of the data management plan and data flows, main web-portal and interoperability protocols.
An important part of the EuCanShare platform is the data access manager, a tool for the data owners to control the access to their sensitive datasets. We built a user-friendly interface for data access committees (data owners) to easily manage their data requests and access credentials. The data access portal facilitates the creation and internal organization of the data access committees as well as the linkage of data usage conditions to specific datasets. The interface includes filters for browsing requests and page to visualize the request history helping with the handling of data access requests.
EUCANCan: toward a federation of clinical institutions in oncology
The EUropean-CAnadian CANcer Network worked towards building a federated network to advance personalized medicine in oncology, by promoting the standard analysis, management and sharing of harmonized genomic and phenotypic data. The EGA led tasks on defining data flows and preparing an adapted infrastructure for long-term data storage and sharing. One of the key contributions was the conception of the EGA communities, offering standard and interoperable solutions for data discovery, processing, and sharing to projects and institutions that would like to manage and share data in the context of the EGA ecosystem.
What's Next?
The participation in projects such as CINECA, EUCANCan, and euCanSHare provide us with experience for future projects. We are happy to enhance data sharing and reuse, vital for advancing clinical and genomic research.
The Discovery Portal is a wonderful proof of concept for the Beacon network. Next, we will finalise both the network and the user interface, and make sure it can be applied to more clinically-centred settings like hospitals.
Most data hosted at the EGA are about cancer research. Currently, we contribute to building tools and infrastructure to empower oncology research in several European projects such as EUCAIM, EUCANIMAGE and EOSC4Cancer. We also do not lose sight of initiatives in the field such as the International Cancer Genome Consortium (ICGC) with the project ARGO, whose data model was adopted in EUCanCan.