There are virtually no research fields that do not benefit from the large-scale genomic analysis that are so popular nowadays. For research into diseases with highly diverse clinical behavior like pediatric neuroblastoma, nevertheless, this type of approach has been a game-changer.
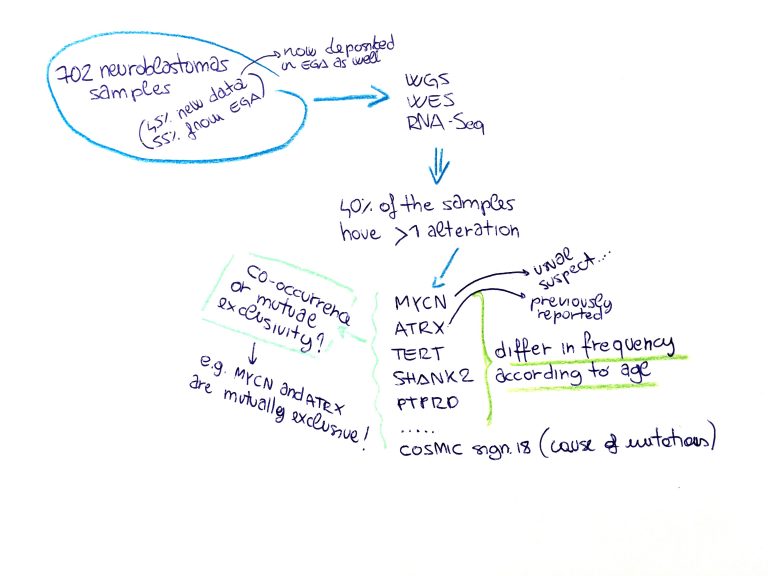
Samuel W. Brady and colleagues, from St. Jude Children’s Research Hospital in Memphis, have gathered data from 702 neuroblastomas which were profiled by whole genome sequencing (WGS, n = 205), whole exome sequencing (WES, n = 539), and/or RNA-Seq. Less than half of the samples were generated during this study, while the others were obtained from previous work. Part of the WES data was indeed deposited in EGA in 2012 (EGA study EGAS00001000213).
“These findings suggest that the sympathetic nervous system, the tissue from which neuroblastoma arises, is susceptible to different oncogenic insults at different times during development, which could be explored in future investigations using animal models.” Conclude the authors, highlighting the importance of their findings.
Pediatric neuroblastomas are classified according to clinical and molecular features into low, intermediate, and high risk. Age plays a critical role in the classification, with younger children having an overall better prognosis. The samples included in this study belonged to all ages and risk-groups: the aim of such set up is the identification of age-associated molecular aberrations.
The authors defined a mutational landscape of pediatric neuroblastoma including rare driver events, impossible to pick with a smaller data set. Interestingly, most driver mutations' frequency was found to be age-related. Those include alterations on MYCN, TERT, PTPRD, and Ras pathway, in addition to the previously reported ATRX. In addition, the large data set allowed the authors to define co-existence relations between the mutations (mutual exclusion or co-occurrence) which is a crucial puzzle piece to unravel the mechanisms driving the pathogenesis.
Enjoy this really interesting article, open source on Pubmed.